Monthly Shaarli
June, 2025
I have a Quarto project of manuscript type with notebooks that generate tables and figures. And I would like to include specific tables from any notebook in the main manuscript index.qmd.
In tackling this question I was constrained by the following factor:
- never
{{< include ... >}}a notebook into the main manuscript, because I found no way to do that without polluting the main manuscript with the entire contents of said notebook. This was unacceptable to me since the whole point of separating analysis into separate files was to keep the main manuscript "clean" and easy to work with for some co-authors who might not need or want to see all the plumbing.
A figure from a notebook is easily included in the main manuscript with {{< embed /notebooks/notebook.qmd#fig-id >}} which renders the plot, its caption and (assuming you defined a label) also numbers it and makes it cross-referable. Unfortunately no such mechanism is offered for tables (I assume because tables are not stand-alone image files that can easily be embedded).
Here is the hack of a solution that I arrived at after wrestling with this today. As far as I know I have not seen this particular approach mentioned anywhere else before.
In the notebook
Create a Markdown table using knitr::kable and save the output (i.e., the rendered Markdown table) to a file on disk, for example:
sas.fityk.params %>%
select(temperature, diameter, starts_with("fit.")) %>%
knitr::kable(col.names = c("$d/\\unit{\\nm}$", "$T_\\text{a}/\\unit{\\degreeCelsius}$",
"SSR", "WSSR", "DoF", "WSSR/DoF", "$R^2$"), digits = c(0, 1, 1, 2, 1, 1, 1, 6),
align = c("l", "l", "l", "l", "l", "l", "l", "l")) %T>%
print() %>%
saveRDS(file = here("assets/objects/tab-sas-fitinfo.rds"))Note that print is necessary if you want to also render the table in the notebook's output.
And since the Markdown will be handled by pandoc we can also use LaTeX math. But another effect of this being Markdown is that the Quarto -> MathJax pipeline is never involved, so more complex LaTeX cannot be used. I wonder if there's a way to force Quarto to handle a particular chunk even if it would normally just be passed on to pandoc...?
In the main manuscript
```{r}
#| results: asis
this.cap <- paste("Goodness of fit parameters for the anti-Stokes/Stokes spectra.")
this.lbl <- "{#tbl-sas-fitinfo}"
cat(c(
readRDS(file = here("assets/objects/tab-sas-fitinfo.rds")),
paste(paste0("\n", ":"), this.cap, this.lbl, "\n")),
sep = "\n")
```This re-produces the same table as in the notebook, and with a numbered table caption that can be cross-referenced. This works in rendered HTML output and even works in PDF (by some magic). I don't care too much about DOCX right now, so that's success!
Avenues that did not work
My first idea was to simply print the Markdown table in a chunk with a tbl-cap:
```{r}
#| results: asis
#| tbl-cap: Goodness of fit parameters for the anti-Stokes/Stokes spectra.
# label: tbl-sas-fitinfo
readRDS(file = here("assets/objects/tab-sas-fitinfo.rds"))
```but this simply results in a Quarto error (apparently triggered by the presence of label):
Error running filter /usr/lib/rstudio-server/bin/quarto/share/filters/main.lua:
Block, list of Blocks, or compatible element expected, got tableThat prompted me to try printing the caption line manually, but that too fails, surprisingly:
```{r}
#| results: asis
this.cap <- paste("Goodness of fit parameters for the anti-Stokes/Stokes spectra.")
this.lbl <- "{#tbl-sas-fitinfo}"
cat(paste(paste0("\n", ":"), this.cap, this.lbl, "\n"))
readRDS(file = here("assets/objects/tab-sas-fitinfo.rds"))
```Turns out that in the intermediate Markdown file created by Quarto, every line in the chunk is printed inside its own fenced block, and thus the caption line becomes syntactically separate from the table itself.
The solution (as shown above) is to print the caption line and the table itself in a single command.
Links
- https://github.com/quarto-dev/quarto-cli/discussions/2052 - How to gracefully mix Markdown and latex tables?
- https://github.com/quarto-dev/quarto-cli/discussions/11067 - How to include pre-rendered tables?
- https://forum.posit.co/t/tables-numbering-in-quarto/196413 - tables numbering in quarto
- https://github.com/quarto-dev/quarto-cli/discussions/9334 - debugging advice for main.lua errors
- https://stackoverflow.com/questions/74097579/make-figure-text-bold-in-quarto-figure-captions - Make "Figure" text bold in quarto figure captions
- https://scholar.social/@solarchemist/114678486113073128
sessionInfo
$ quarto check # slightly cleaned-up output
Quarto 1.5.57
[✓] Checking versions of quarto binary dependencies...
Pandoc version 3.2.0: OK
Dart Sass version 1.70.0: OK
Deno version 1.41.0: OK
Typst version 0.11.0: OK
[✓] Checking versions of quarto dependencies......OK
[✓] Checking Quarto installation......OK
Version: 1.5.57
Path: /usr/lib/rstudio-server/bin/quarto/bin
[✓] Checking tools....................OK
TinyTeX: (not installed)
Chromium: (not installed)
[✓] Checking LaTeX....................OK
Using: Installation From Path
Path: /usr/local/texlive/2023/bin/x86_64-linux
Version: 2023
[✓] Checking basic markdown render....OK
[✓] Checking Python 3 installation....OK
Version: 3.10.12
Path: /usr/bin/python3
Jupyter: (None)
[✓] Checking R installation...........OK
Version: 4.5.0
Path: /opt/R/4.5.0/lib/R
knitr: 1.50
rmarkdown: 2.29
[✓] Checking Knitr engine render......OKReduction-oxidation sum reactions are typically written as two lines (red/ox) with a sum reaction underneath. Here is an example of how to typeset this building on the reactions environment from the awesome chemmacros package.
My goals with this custom environment was to retain chemical reaction numbering (with sub-numbering for the redox reactions) and horizontally align everything on the reaction arrow.
The solution is not automated all the way so may require some fiddling of horizontal or vertical distances to make it look great.
Load chemmacros in the preamble:
\usepackage[
% *minimal* specifies the following set of always loaded modules:
% acid-base, charges, nomenclature, particles, phases, symbols
minimal=true,%
modules={reactions}%
]{chemmacros}and define the custom environment I like to call subreactions:
\makeatletter
\newenvironment{subreactions}{%
\refstepcounter{reaction}%
\protected@edef\theparentequation{\thereaction}%
\setcounter{parentequation}{\value{reaction}}%
\setcounter{reaction}{0}%
\def\thereaction{\theparentequation\alph{reaction}}%
\ignorespaces
}{%
\setcounter{reaction}{\value{parentequation}}%
\ignorespacesafterend
}
\makeatotherI did not come up with that on my own, I got a lot of help and found inspiration in the work of others.
Here is the end result (screenshot from my thesis), the well-known water splitting reaction expressed as hydrogen reduction and water oxidation:
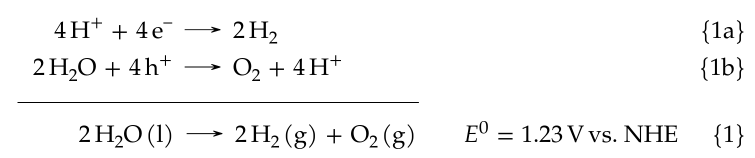
which was created with this code:
\begin{subreactions}\begin{reactions}%
4 \proton{} + 4 \electron{} &-> 2 \hydrogen{} && $\qquad\qquad\qquad\qquad$ \AddRxnDesc{Hydrogen~evolution} \"\label{rxn:hydrogen-evolution}\" \\%
2 \water{} + 4 \hole{} &-> \oxygen{} + 4 \proton{} && $\qquad\qquad\qquad\qquad$ \AddRxnDesc{Oxygen~evolution} \"\label{rxn:oxygen-evolution}\"%
\end{reactions}\end{subreactions}%
\addtocounter{reaction}{-1}%
\vspace{-\baselineskip}%
% note that the following lengths must be adjusted if
% the horizontal extent of any reactions are changed!
\hspace{13mm}%
\begin{minipage}{59mm}%
\vspace{-\baselineskip}%
\hrulefill%
\end{minipage}%
\begin{reaction}%
\qquad{}\qquad{}\quad{} 2 \water\lqd{} -> 2 \hydrogen\gas{} + \oxygen\gas{} $\quad\enspace\enspace E^0=\qty{1.23}{\voltNHE}$ \AddRxnDesc{Overall~water~splitting} \"\label{rxn:water-splitting}\"%
\end{reaction}%